Hi,
I'm trying to use the RT_MOTIF simulation parameter to introduce sequence bias into the reverse transcription phase of a Flux Simulator library construction and sequencing run. I've set the parameter to the value "default", which, if I understand correctly, should use a PWM derived from an Illumina protocol to bias the positions in each fragment at which priming events occur during reverse transcription.
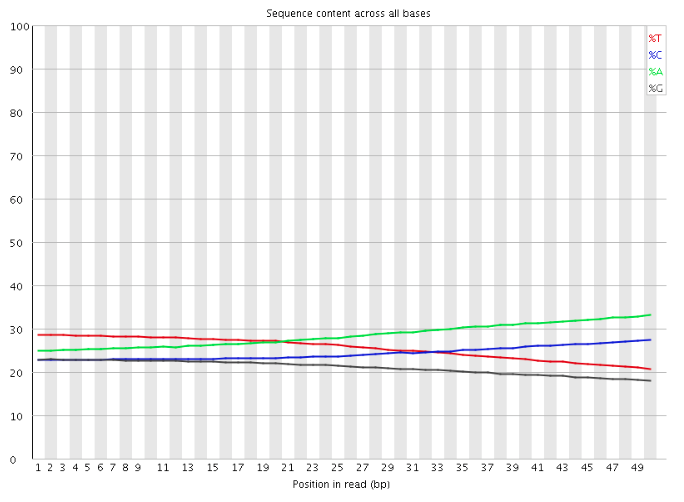
Where I am confused is that I would have then expected the sequence pattern encoded in the default PWM to be evident upon examination of the reads that are output at the end of the Flux Simulator run, for example using a tool such as FastQC. If I use FastQC to examine the reads produced by a simulation run with RT_MOTIF set to "default", I see something like the following for the "Per base sequence content" graph:
where I was instead expecting to see a pattern of bias in the bases at the start of the reads, reflecting the sequence pattern encoded in the default PWM (and, indeed, a graph of "Per base sequence content" for a Flux Simulator run in which the RT_MOTIF parameter is left at its default value of none looks very similar).
That is, I do not appear to be obtaining any sequence bias in my reads when using the RT_MOTIF parameter - am I misunderstanding its function or how to use it? I'm using Flux-Simulator v1.2.1 (Flux Library: 1.22).