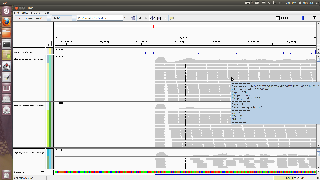
Hello
I got a strong 5' bias. First positions in a transcript have a high( very high coverage) . The window that follows has almost no reads.
I believe the figure should speak for itself. I join also the parameters file used.
Any suggestions on how to reduce this bias?
Thank you,
Bogdan
PARAMETER TABLE:
NB_MOLECULES 50000
TSS_MEAN NaN
POLYA_SCALE 50
POLYA_SHAPE 2
## Fragmentation
FRAG_SUBSTRATE RNA
FRAG_METHOD UR
FRAG_NB_LAMBDA 50
## RT parameters
RTRANSCRIPTION YES
RT_PRIMER RH
RT_LOSSLESS YES
RT_MIN 2000
RT_MAX 6000
## PCR / Filtering
## no [GC] bias to not observe sequence based biases
PCR_DISTRIBUTION default
PCR_PROBABILITY 0.5
GC_MEAN NaN
FILTERING YES
SIZE_SAMPLING MH
# Sequencing
FASTA true
READ_NUMBER 1000000
READ_LENGTH 50
PAIRED_END NO